Chemists at the University of Illinois Urbana-Champaign (UIUC) have made a breakthrough in understanding the crystal structure of a crucial component of the monensin enzyme. This discovery has led to the unlocking of the enzyme’s reaction activity mechanism. The research, published in Nature Communications, was a collaborative effort between UIUC and The University of Texas at El Paso (UTEP). In this article, we will delve into the details of this groundbreaking study and its potential implications for the future of antibiotic design.
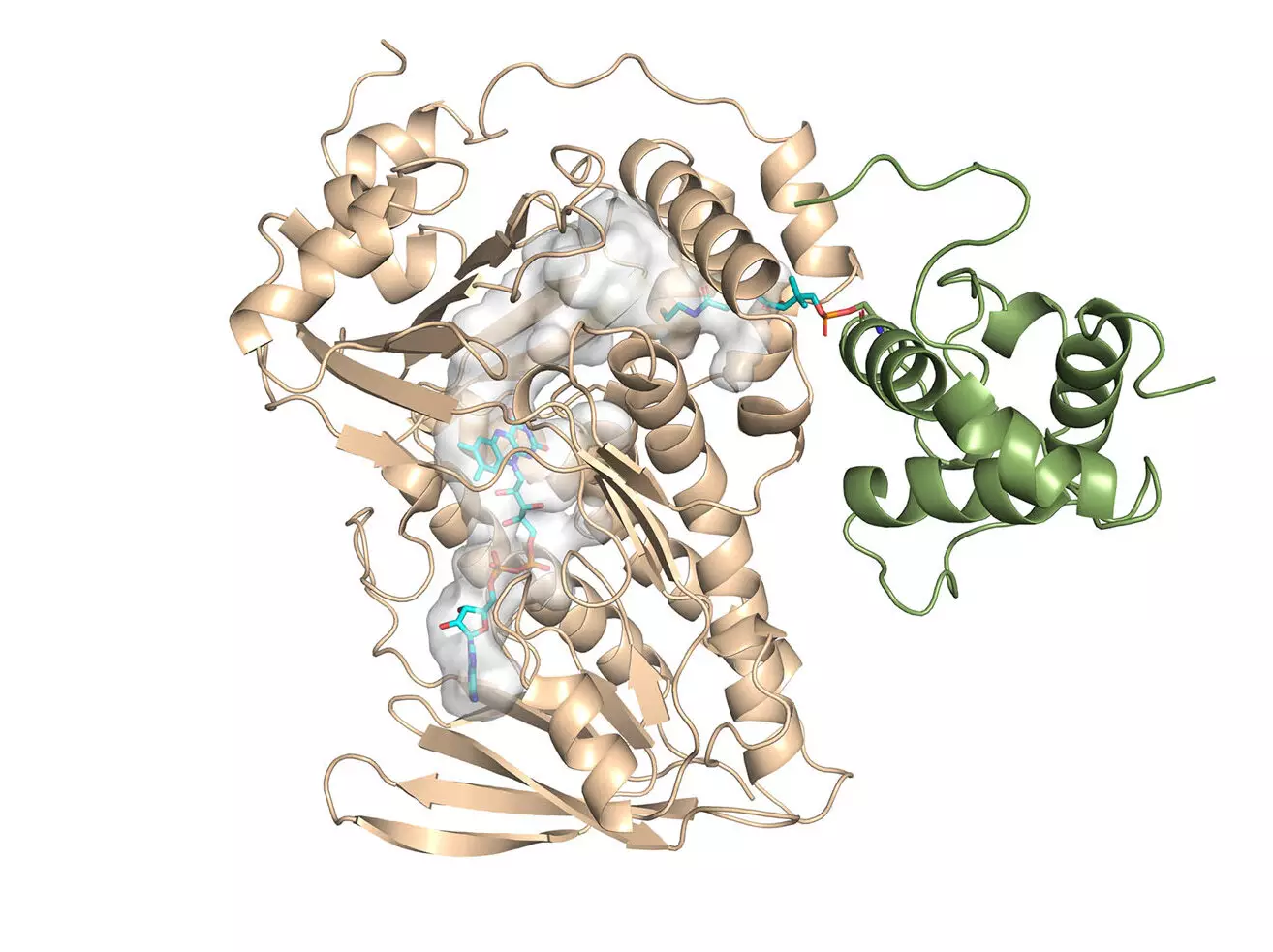
The primary achievement of this research was the determination of the crystal structure for MonCI, the key enzyme responsible for synthesizing monensin in soil bacteria. Led by Professor Chu-Young Kim from UIUC, the experimental team successfully solved the crystal structure of MonCI. This significant finding provides a foundation for further investigations and potential engineering of new antibiotics.
The computational aspect of the study was conducted by Associate Professor Lela Vukovic from UTEP. Her team utilized the computational power of the Lonestar6 system at the Texas Advanced Computing Center (TACC) to analyze the monensin research. Supported by the University of Texas Research Cyberinfrastructure (UTRC) initiative, Vukovic’s team performed simulations to model the enzyme and its substrate. These simulations offered valuable insights into the reaction sequence that produces monensin.
By understanding the mechanism of MonCI, the researchers have paved the way for designing safer and more effective antibiotics. The study revealed that MonCI carries out three crucial epoxidation reactions, which is a rare occurrence. This finding has profound implications for engineering bacteria to produce new antibiotics. By manipulating the enzyme’s structure and reaction sequence, scientists can potentially develop improved versions of monensin that are less toxic to animals.
Performing computational studies on complex biological molecules like MonCI and monensin is no easy task. However, Vukovic’s team overcame these challenges with the help of supercomputers, particularly the NAMD software developed by the late Klaus Schulten’s group at UIUC. This software allowed them to zoom into the enzyme and analyze the sequence of reactions that generate monensin. The optimization of NAMD for running on supercomputers like Lonestar6 and Stampede2 at TACC highlights UIUC’s commitment to advanced computational research.
While this study focused on MonCI, it is only one component of the monensin biosynthesis process. The research team emphasizes the need for further investigation into the other enzymes involved. Their ultimate goal is to generate improved versions of monensin that can better serve the agricultural industry without posing risks to animals. Currently, monensin is toxic to horses and dogs, causing accidental poisoning and death. A nontoxic alternative is urgently required to address these concerns.
The crystal structure revelation of MonCI, a vital component of the monensin enzyme, marks a significant milestone in our understanding of its reaction activity. Through a combination of experimental and computational studies, researchers were able to unravel the complex mechanisms involved in monensin synthesis. This breakthrough opens up new opportunities for antibiotic design and paves the way for safer and more effective antibacterial agents. With ongoing advancements in computational research and the continuous optimization of software like NAMD, the future looks promising for the development of innovative solutions in the field of biochemistry.

Leave a Reply