Understanding the intricate interactions between proteins within cells is crucial for deciphering the complex molecular machinery that drives various biological processes. However, our current knowledge of protein-protein interactions often lacks cellular contexts, as most studies are conducted in artificial systems or isolated cells. This limitation hinders our ability to comprehensively understand how proteins function in living organisms. In a recent study, a research team from The University of Hong Kong (HKU) developed a novel approach to investigate tissue-specific protein interactions, shedding light on the importance of such research in advancing our understanding of cellular processes.
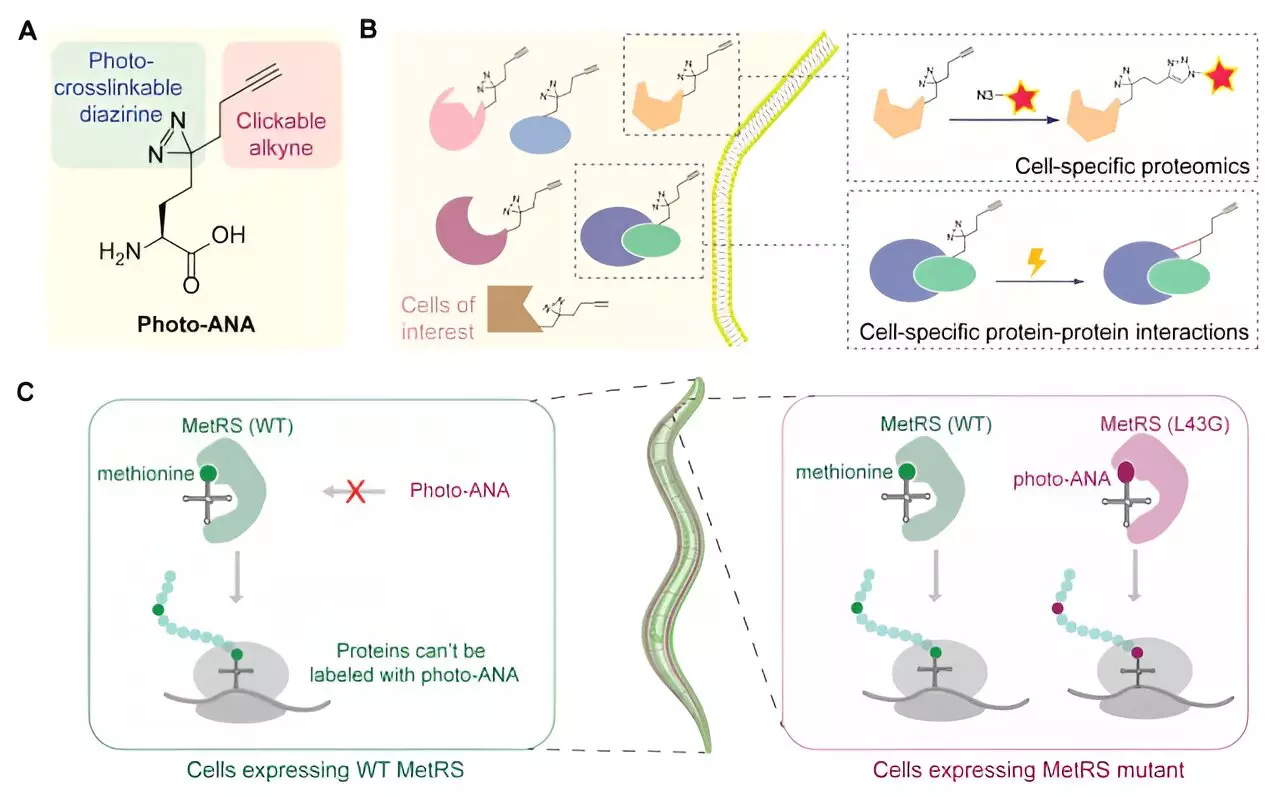
The collaborative team led by Professor Xiang David Li and Professor Chaogu Zheng from the Department of Chemistry and the School of Biological Sciences at HKU, respectively, introduced the Methionine Analog-based Cell-Specific Proteomics and Interactomics (MACSPI) method. This innovative chemical biology approach involves labeling proteins from specific cells with a bifunctional amino acid probe, allowing for the isolation and capture of protein-protein interactions through photo-crosslinking.
The team designed and synthesized an unnatural amino acid, photo-ANA, that structurally resembles methionine but contains additional components. The alkyne group serves as a chemical handle for the extraction and purification of labeled proteins, while the diazirine group is activated by light to form stable covalent linkages with interacting molecules. By engineering the enzyme MetRS to incorporate the unnatural amino acid into proteins, the team achieved tissue-specific labeling of proteins, enabling the capture and isolation of protein complexes from specific tissues.
As a proof-of-concept, the researchers applied the MACSPI method to profile proteins from muscle cells and neurons in the model organism C. elegans. This led to the discovery of numerous novel tissue-specific proteins and interactions. Additionally, the team demonstrated the method’s utility by identifying tissue-specific interactors of the ubiquitously expressed protein HSP90, revealing distinct protein binding patterns in muscles and neurons that regulate different biological processes.
By enabling the profiling of proteomes and interactomes with spatial and temporal specificity, the MACSPI method holds great promise for advancing biological and biomedical research. Understanding tissue-specific protein interactions not only enhances our knowledge of cellular processes but also provides valuable insights into disease mechanisms. For instance, the identification of neuronal HSP90 interactors associated with neurodegeneration in a Parkinson’s disease model showcases the potential of this approach in uncovering novel therapeutic targets.
The development of innovative methods like MACSPI highlights the importance of investigating tissue-specific protein interactions in enhancing our understanding of cellular functions. By bridging the gap between in vitro and in vivo studies, this approach opens up new possibilities for studying complex biological systems and addressing challenging research questions in various fields. The team’s groundbreaking work underscores the significance of integrating chemical biology, proteomics, and cell biology to unravel the mysteries of protein-protein interactions in living organisms.

Leave a Reply