Recent advancements in the study of protein synthesis have emerged from the University of Tsukuba, where researchers have successfully created a sophisticated model that closely simulates the ribosomal environment. This innovative approach offers a significant leap forward in our understanding of how ribosomes—cellular structures responsible for protein formation—create complex proteins. By harnessing computer simulations to delve deeper into the internal workings of ribosomes, this research sheds light on previously obscure mechanisms, particularly those involving the ribosomal tunnel through which nascent proteins exit.
In the realm of cellular biology, the ribosome tunnel has often been viewed as a mere passageway for protein release. However, emerging studies have revealed the tunnel’s far more significant role in the initial stages of protein folding. Evidence suggests that many proteins begin to adopt their crucial three-dimensional structures while still traversing this tunnel. Despite the existence of these observations, the underlying biochemical interactions and properties that influence this process have been elusive. This research endeavors to bridge that gap by providing comprehensive 3D analyses of ribosome tunnels.
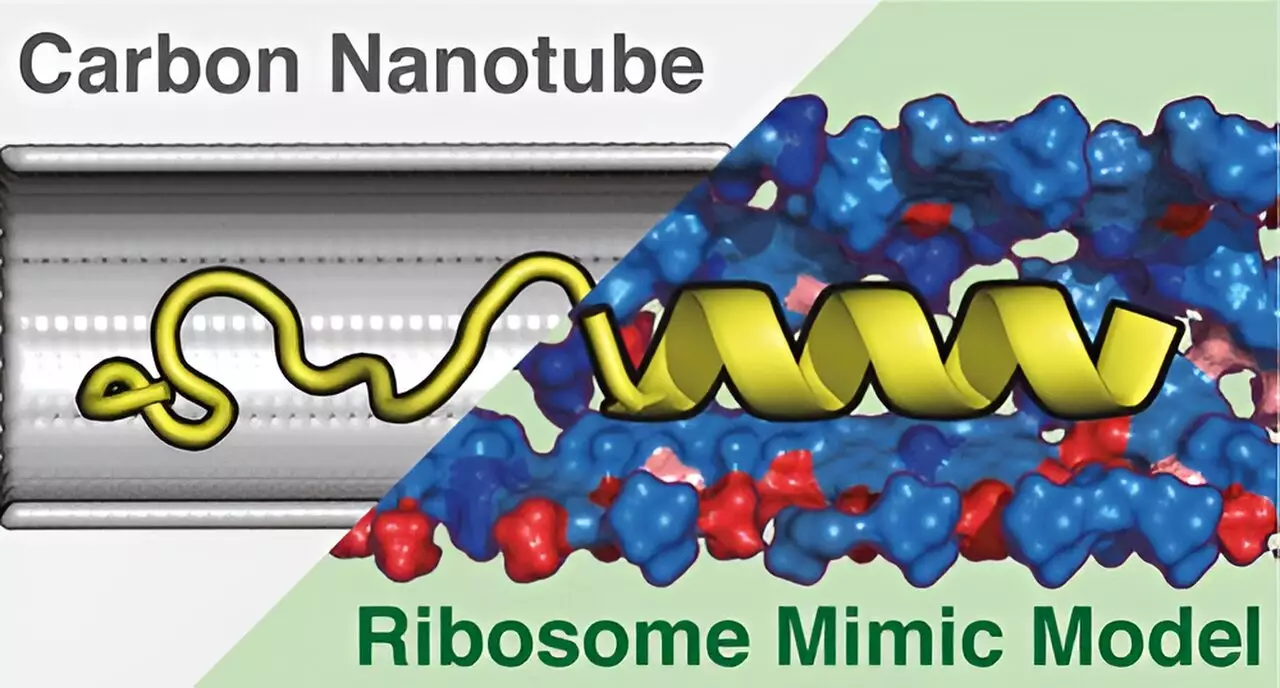
To tackle the complexities surrounding the structure and function of ribosomes, the research team developed a novel cylindrical structure dubbed the Ribosome Environment Mimicking Model (REMM). This model meticulously replicates both the inner diameters of the ribosomal tunnel and its specific chemical properties. Most importantly, REMM does not merely recreate the tunnel’s shape but also incorporates the chemical environment essential for accurate protein folding. Such characteristics are pivotal in understanding how proteins attain their spatial configurations and functional capabilities.
In contrast, the team also created a Conventional Carbon Nanotube (CNT) model, which depicted only the geometric dimensions of the tunnel, entirely omitting the chemical context. This distinction is crucial, as it illuminates the fundamental limitations present in oversimplified models that can hinder scientific progress in this field.
Utilizing molecular dynamics simulations, the researchers meticulously examined the protein structures within both REMM and CNT frameworks. Findings from these simulations disclosed a remarkable divergence in their efficacy: the REMM remarkably matched the experimentally observed protein structures, while the CNT model fell short. This disparity underscores the importance of the chemical environment alongside geometric considerations in protein synthesis.
Furthermore, the investigation uncovered that the chemical diversity within the REMM is a vital factor contributing to its superior predictive capabilities. The understanding gleaned from this study paves the way for enhanced exploration of protein conformations within live cells, potentially catalyzing breakthroughs in biomedical research and therapeutic approaches.
As scientists continue to refine and develop the REMM, the implications extend well beyond the academic realm. The insights derived from this innovative model could impact various fields, from drug design to biotechnology, where a refined understanding of protein behavior is essential. Though challenges remain, the progress made by the University of Tsukuba represents a pivotal moment in molecular biology, promising to advance our comprehension of the intricate processes governing life at a cellular level.

Leave a Reply