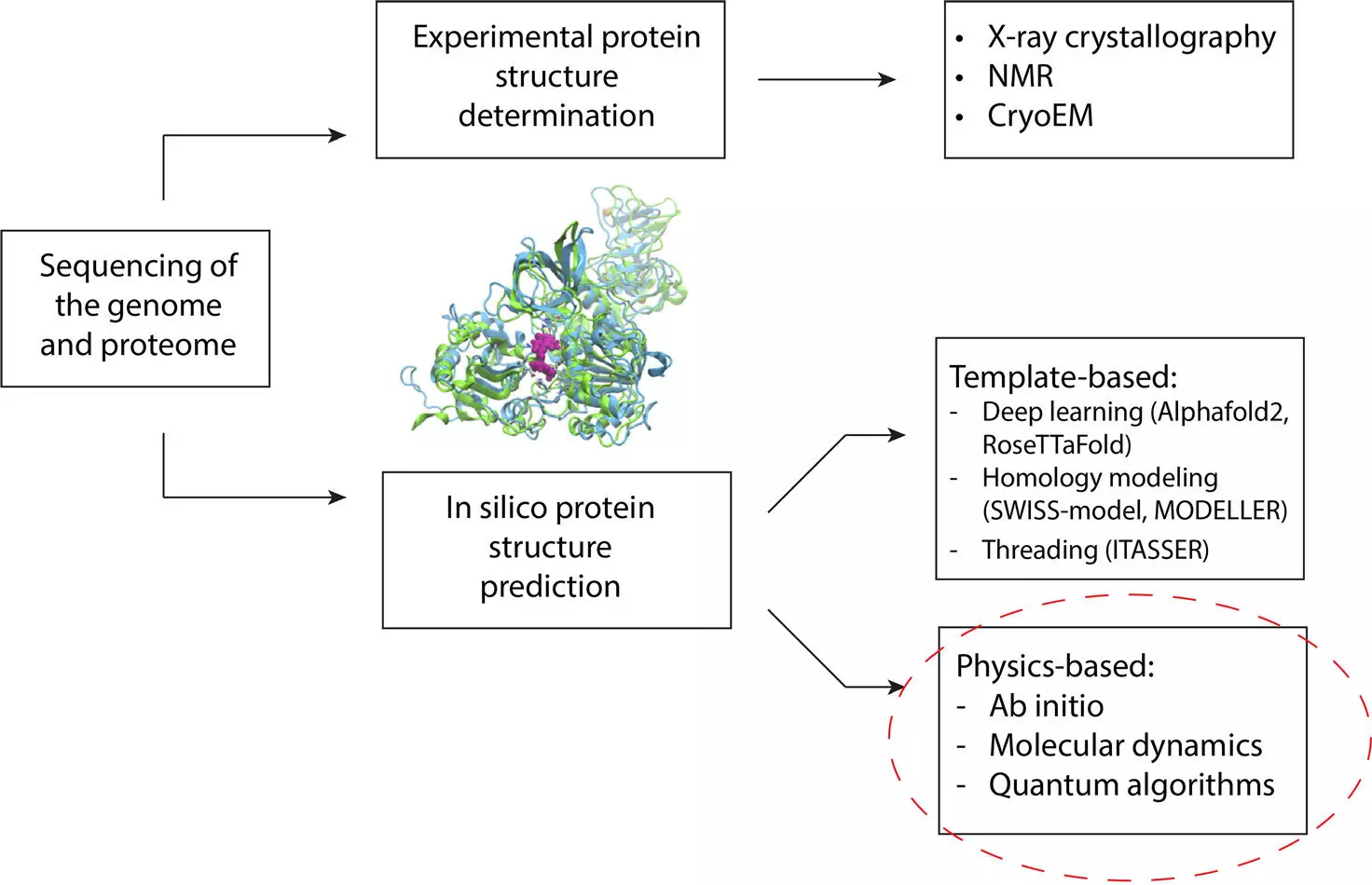
Protein structure prediction has been a key area of focus for researchers due to its implications on human health and disease. Computational approaches have long been utilized to predict how proteins fold themselves into structures that determine their functions and interactions with other molecules in the body. These structures play a crucial role in understanding disease mechanisms and developing effective therapies. However, traditional methods have limitations when it comes to predicting the structures of mutated or novel proteins.
Recent findings by researchers from Cleveland Clinic and IBM have shed light on how quantum computing can revolutionize protein structure prediction. By combining quantum and classical computing methods, the research team was able to overcome the limitations faced by traditional machine learning techniques. Quantum algorithms were used to model the lowest energy conformation of protein fragments, while classical approaches were employed for final structure refinement. This hybrid framework outperformed traditional methods, including the state-of-the-art AlphaFold2, showcasing the potential of quantum computing in this field.
The research team’s work marks a significant advancement in the field of protein structure prediction. By leveraging the power of quantum computing, they were able to accurately predict the folding of a Zika virus protein fragment, demonstrating the framework’s capabilities in handling complex protein structures. The use of quantum algorithms to address computationally demanding steps in the prediction process highlights the potential for more accurate and efficient models.
One of the key strengths of this project lies in the interdisciplinary collaboration among experts from various fields. The team’s diverse expertise, spanning computational biology, chemistry, structural biology, software engineering, and quantum computing, was instrumental in developing a comprehensive computational framework. By deconstructing the prediction process into quantum and classical components, the researchers were able to achieve unprecedented accuracy in modeling protein structures.
The successful application of quantum computing in protein structure prediction opens up a world of possibilities for advancing our understanding of complex biological processes. The research team plans to further optimize quantum algorithms to predict the structures of larger and more intricate proteins. This ongoing work represents a crucial step forward in exploring the potential of quantum computing in biomedical research. Dr. Hakan Doga emphasizes the importance of this breakthrough in paving the way for new advancements in protein structure prediction.
The integration of quantum computing methods in protein structure prediction represents a groundbreaking achievement that holds tremendous promise for the future of biomedical research. By combining the strengths of quantum and classical computing, researchers have unlocked new possibilities in accurately modeling protein structures. This innovative approach could potentially revolutionize drug design, disease prevention, and our understanding of complex biological systems. The collaborative effort of experts from diverse disciplines has paved the way for a new era in computational biology and quantum algorithm design. The future of protein structure prediction looks brighter than ever, thanks to the pioneering work of the research team from Cleveland Clinic and IBM.

Leave a Reply